Table of Contents
Gene Regulation and Eukaryotic Gene Structure
Because DNA is found in the cytoplasm of prokaryotes, translation can begin before transcription is completed. As a result, the majority of gene regulation in prokaryotes happens at the transcriptional level. In eukaryotes, however, the nuclear envelope separates the DNA from the cytoplasm, thus mRNA must go from the nucleus to the ribosomes.
As a result, gene activity can be controlled during transcription, translation, or even post-translational modification of the translated protein. Eukaryotic genes are not grouped into operons regulated by a single promoter, which is another distinction. The majority of genes only code for one protein, and each gene has its own promoter.
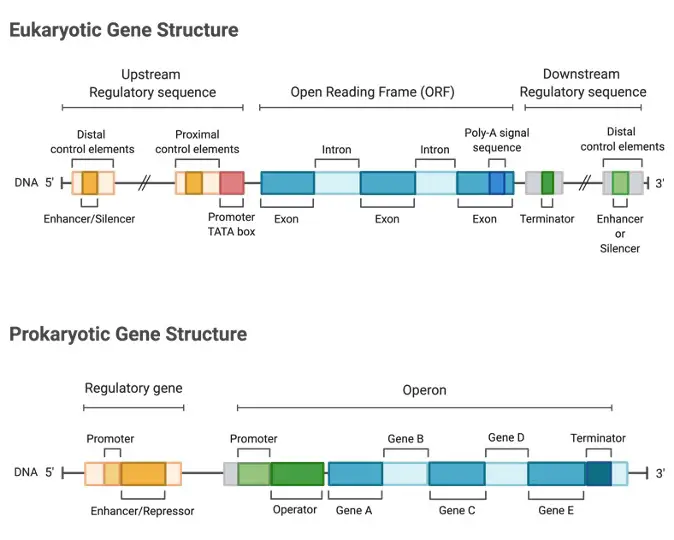
In most protein-coding genes, the coding sequence is split up into multiple exons separated by non-coding introns rather than being continuous. To generate a translatable mRNA, the introns are deleted after transcription. A core promoter that binds RNA polymerase and other surrounding areas that bind other regulators are seen in eukaryotic promoters.
The three DNA binding areas depicted in the image: the TATAA box in the core promoter and the CAAT and GC boxes in the proximal promoter, may or may not be present in eukaryotic promoters.
Gene Regulation and Transcription Factors
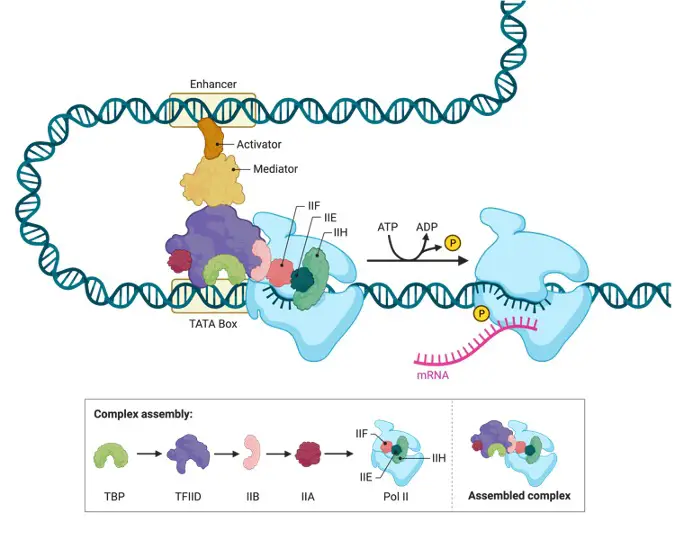
In eukaryotes, two kinds of protein transcription factors control transcription. With the help of basal transcription factors, RNA polymerase attaches to the core promoter, one of which is a TATA-binding protein that recognises the TATAA region of the core promoter DNA.

Additional areas adjacent to the main promoter bind particular transcription factors. To increase or inhibit transcription, certain transcription factors bound to the enhancer region interact with those linked to the promoter. Transcription factors tied to the enhancer region are only active when the enhancer DNA is linked to them.
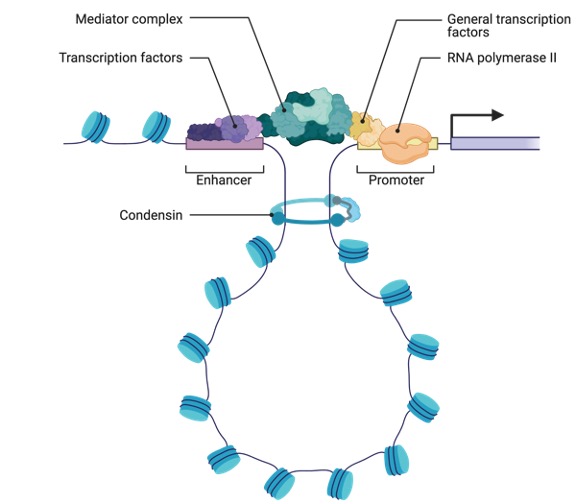
Because the enhancer region can be hundreds or even thousands of nucleotides upstream of the promoter they control, the DNA must bend to allow the enhancer-bound proteins to interact with the promoter-bound proteins.
Because of the nature of eukaryotic chromosomes, another layer of control is required in eukaryotes. While prokaryotic chromosomes are made up of single, typically circular DNA molecules, chromatin is made up of DNA and proteins in eukaryotic chromosomes. Histones are a kind of chromosomal protein that creates protein complexes around which DNA is wrapped to form nucleosome structures.
Most eukaryotic genes are not easily accessible for transcription as these nucleosome complexes become increasingly condensed with other proteins. The opening up of the chromosome to allow different transcription factors and RNA polymerase access to the DNA is the initial stage of eukaryotic gene control.
Post-Transcriptional Regulation
Once a eukaryotic gene has been transcribed, the introns must be removed before it can be processed. Alternative splicing, or the selection of various groups of exons to create the mRNA, can produce multiple different copies of the protein coding sequence in bigger genes with many exons.
Furthermore, nucleotides are added to both the 5′ and 3′ ends of the mRNA to protect it from destruction in the cytoplasm. A methyl-guanosine cap is found at the 5′ end of the mRNA, which helps the ribosome recognise it. The poly-A tail is a string of adenine nucleotides at the 3′ end of the mRNA that serves to stabilise it as it travels to the ribosome.

Proteins that bind to the untranslated 5′ and 3′ regions of the mRNA can then either promote or suppress mRNA translation. In reaction to different variables in the cell’s environment, these proteins may be altered. Regulatory RNAs may attach to an mRNA to influence its translation in addition to these regulatory proteins.
The protein can be further changed once it has been translated. The starting methionine, which is encoded by the start codon AUG, is frequently deleted. Trypsin and other digestive enzymes, for example, are produced as an inactive precursor protein that must be activated by removing a portion of the protein.
Insulin’s protein precursor is a bigger protein that is broken into multiple pieces, two of which are then joined together to form active insulin’s A and B chains. Two separate genes produce the alpha and beta chains of the protein haemoglobin. In general, transcriptional regulators control which genes are transcribed in a cell, whereas post-transcriptional factors govern whether the encoded protein product is produced.
MyoD and Muscle Cell Differentiation
MyoD is a transcription factor that can cause muscle tissue to differentiate. It’s just one of many regulators involved in muscle development, but it’s plenty to show how complicated eukaryotic gene control can be. Skeletal muscle is a multinucleate tissue made up of embryonic myoblasts that have fused together.
MyoD, Myf5, Myogenin, and MRF4 are four key regulators of muscle cell development. Mice lacking either MyoD or Myf5 create normal muscle tissue, whereas mice lacking both MyoD and Myf5 fail to generate any skeletal muscle at all, indicating that these two genes regulate muscle development jointly.
Mice lacking the other two genes, on the other hand, have a variety of muscular impairments, indicating that all four genes are necessary. MyoD binds to the regulatory areas of several of the genes involved in skeletal muscle formation.
It generally does not operate alone, but rather forms heterodimers with other proteins that have comparable functions. These dimers detect pairs of E-box DNA regulatory elements, the consensus sequence of which is CANNTG.
MyoD may also play a role in muscle cell differentiation by attaching to an enzyme that acetylates histones, which allows it to rearrange the chromosomes around muscle-specific genes, making them accessible to transcription. Histones are attracted to the negatively charged phosphate groups of the DNA backbone because they are basic proteins.
Acetylating histones lowers their affinity for DNA, allowing it to be released for transcription. The functions of MyoD in activating muscle-specific genes are oversimplified, and MyoD itself has many regulators. It is, however, a brief glimpse into the intricacies of eukaryotic transcriptional control.
Gene Regulation Citations
- Enhancer DNA methylation: implications for gene regulation. Essays Biochem . 2019 Dec 20;63(6):707-715.
- Modulating gene regulation to treat genetic disorders. Nat Rev Drug Discov . 2020 Nov;19(11):757-775.
- Gene Regulation in and out of Equilibrium. Annu Rev Biophys . 2020 May 6;49:199-226.
- Figures are created with BioRender.com







