Table of Contents
Nucleobase Definition
Nucleobase refer to the nitrogenous bases that are a part of nucleotides that are monomers of nucleic acid. For example, purines.
What is Nucleobase?
Nucleobase comprised by monomeric units nucleotides, DNA is a biopolymer organic compound. A nucleotide consists of a 5-Carbon sugar, base, and phosphoric acid. The nucleotides form 3′, 5′ phosphodiester linkages that link them to each other.
For the formation of this bond, a 5′-phosphoric group of a nucleotide is esterified with the 3′-hydroxyl of the next monomeric nucleotide. Bases are components of nucleosides and also contain nitrogen. They attach to a pentose sugar via a glycosidic bond to form nucleosides. The sugar can be deoxyribose or ribose depending upon the nucleic acid.
Nucleosides further form nucleotides when phosphoric acid is attached. The nucleobase is paired in the case of double-stranded DNA, where they pair in a complementary fashion.
Nucleobase Classification
The bases belong to 2 categories, purines, and pyrimidines. These 2 types differ in respect to their chemical structure and they both constitute aromatic heterocyclic compounds. The pyrimidines consist of 1 carbon pyrimidine ring while the purines have 2 carbon rings in their structure as the pyrimidine ring is conjoined with an imidazole ring.
While both purines and pyrimidines are heterocyclic aromatic compounds, they can be differed from each other based on the chemical structure. The purines occur as two carbon rings whereas the pyrimidines occur as one carbon ring. The pyrimidine base has 2 nitrogen atoms while purine has 4.
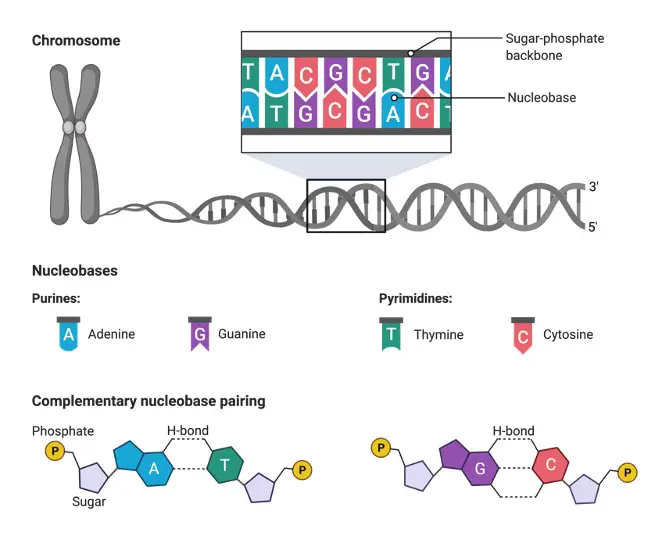
The bases guanine and adenine comprise purine bases while, the bases cytosine, thymine, and uracil comes under pyrimidine. These comprise the 5 canonical primary nitrogenous bases as they decide the sequence of DNA or genes.
DNA and RNA nucleic acid differ in the bases they contain. Unlike in DNA, in RNA uracil pairs with adenine and thymine is not present. Thymine and Uracil both are purines and they differ structurally with respect to a methyl group.
Canonical Nucleobases
The primary nucleobases that comprise the genetic code include cytosine (C), adenine (A), thymine (T), guanine (G), and uracil (U). DNA lacks a uracil nitrogenous base and it has deoxyribose sugar as its constituent. RNA and DNA can be differentiated in the presence or absence of thymine.
Thymine and uracil pair complementary with base Adenine. While cytosine pairs with guanine via 3 hydrogen bonds. The nucleobases that pair with each other are called base complements. DNA lacks uracil in its structures as one of the bases. This may be to prevent instability and efficient functioning of repair systems.
Since the repair system cannot distinguish between deaminated cytosine and original uracil. The presence of the methyl group in thymine prevents this from happening and maintains the integrity of the DNA structure.
Non-primary Nucleobases
Besides the canonical bases, the nucleotides may also comprise other bases like 5-methylcytosine (m5C), which is a pyrimidine base. Other examples of non-primary pyrimidine bases include 5-hydroxymethyl and cytosine 5,6-dihydrouracil. Non-canonical purine bases are xanthine, hypoxanthine, and 7-Methylguanine.
These non-primary bases may be produced by the modification of an existing base. For example, 5-Methylcytosine is actually a methylated cytosine. They may also comprise intermediates like Dihydrouridine that is found as an intermediate in uracil catabolism. Modified adenine may produce non-primary bases like Inosine and Hypoxanthine.
Modification of guanine base can produce Xanthosine and 7-Methylguanosine. The most usually seen non-canonical nucleobase is m5C in the case of DNA. The others like D, Ψ, I, and m7G are seen in RNA especially in tRNA.
Deamination of adenine can lead to the formation of hypoxanthine and xanthine is formed due to the deamination of guanine. Hypoxanthine can also produce xanthine by catalysis of xanthine oxidoreductase and xanthosine can be generated in a reaction catalyzed by purine nucleoside phosphorylase.
Mutagens can enhance the formation rates of xanthine and hypoxanthine. Hypoxanthine is similar in structure to adenine due to which can form faulty base pair with cytosine. The DNA repair systems repair this by base excision repair that requires the enzyme N-methylpurine glycosylase.
Artificial Nucleobases
Bases can also be artificially synthesised. Isocytosine and isoguanine are some examples of nucleobase analogs. Oxidative damage to DNA can lead to the formation of isoguanine that is an isomer of guanine that is employed in hachimoji nucleic acids. Isocytosine can be artificially produced by malic acid and guanidine is an isomer of cytosine and also has applications in hachimoji RNA.
Biosynthesis of Nucleobases
When bases are linked to ribose 5-phosphate they form ribonucleotides that are precursors to bases. Purines are derived from the inosine monophosphate (IMP) nucleotide that is produced from ribose phosphate. Pyrimidines are biosynthesized via a de novo synthesis pathway. In this pathway, the pyrimidine ring is formed first and the pathway results ultimately in the production of UMP.
The major 4 steps include:
- Synthesis of carbamoyl aspartate
- Formation of dihydroorotate
- Ribose phosphate moiety is added
- Formation of UMP
Through this pathway TTP, UTP, and CTP are formed. Pyrimidines are ultimately degraded into water, CO2, and urea. Cytosine is degraded into uracil that is in turn catabolized into N-carbamoyl- β-alanine that is then converted into β-alanine. At the very end of cytosine metabolism, ammonia and CO2 are released. Thymine is degraded into β-aminoisobutyrate that then enters Kreb’s cycle.
Purines are ultimately degraded into uric acid. In the guanine degradation xanthine is an intermediate. Adenosine is first converted into inosine that is then degraded into hypoxanthine that in turn gets converted into xanthine and finally into uric acid.
Purines can also be recycled using salvage pathways. APRT enzyme helps in recycling adenine, while HGPRT enzyme helps in the salvage pathway of guanine and hypoxanthine.
Biological Importance of Nucleobase
The nucleobases are the monomers of the structure of nucleic acid and comprise the genetic code.
Nucleobase Citations
- Functional Systems Derived from Nucleobase Self-assembly. ChemistryOpen . 2020 Apr 1;9(4):409-430.
- Supramolecular nucleobase-functionalized polymers: synthesis and potential biological applications. J Mater Chem B . 2020 Feb 26;8(8):1576-1588.
- Reversal of nucleobase methylation by dioxygenases. Nat Chem Biol . 2020 Nov;16(11):1160-1169.
- Figures are created with BioRender.com